Statistics [29]: Factor and Principle Component Analysis
Published:
In practice, data may contain many variables, however, not all of them have significant influence on the results we want to analysis or predict. Factor analysis and principle component analysis are two basic methods that aim to reduce the dimension of the data to make it easier to understand and analyze.
Factor Analysis
Factor Analysis is a method for modeling observed variables, and their covariance structure, in terms of a smaller number of underlying unobservable (latent) “factors.”
Notations
Observabale variable (with traits):
Common factors (with factors):
The factor model can be thought of as a series of multiple regressions:
where are called factor loadings.
In matrix form:
where
Assumptions
Mean
Hence,
Variance
, where
is the specific variance.
Correlation
Model Properties
where is called the communality of the variable
.
In matrix form:
where
As is a symmetric matrix, there are
parameters in total, which are to be approximated by
parameters.
Principle Component Method
Let denote the samples, with
denotes the sample variance matrix,
We have eigenvalues and
eigenvectors for this matrix.
Eigenvalues:
Eigenvectors:
Then we can express the covariance matrix with eigenvalues and eigenfactors as
The idea behind the principal component method is to approximate this expression. Instead of summing from to
, we sum from 1 to
, ignoring the
terms.
We can rewrite the covariance matrix as
This yields the estimator for the factor loadings:
To estimate the specific variances , we only need to estimate the diagonal elements,
Maximum Likelihood Estimation Method
Using the Maximum Likelihood Estimation Method, we must assume that the data are independently sampled from a multivariate normal distribution with mean vector and covariance matrix of the form:
The maximum likelihood estimator for the mean vector , the factor loading matrix
and the specific variance
are obtained by finding
,
and
that maximize the maximize the log likelihood given by:
which can be obtained using the multivariate normal distribution given in this post.
Goodness of Fit
To assess goodness-of-fit, we use the Bartlett-Corrected Likelihood Ratio Test Statistic (link):
In the numerator we have the determinant of the fitted factor model for the variance-covariance matrix, and below, we have a sample estimate of the variance-covariance matrix assuming no structure where:
and is the sample covariance matrix.
If the factor model fits well then these two determinants should be about the same and you will get a small value for . Otherwise,
would be large.
Under the null hypothesis that the factor model adequately describes the relationships among the variables, there is
where the degrees of freedom are the difference of unique parameters in the two models.
Factor Rotations
The problem with the analysis above is that some of the variables are highlighted by more than one factors, and some even indicate contradictory results. This does not provide a very clean, simple interpretation of the data. Ideally, each variable would appear as a significant contributer in one column (factor). This is the purpose of factor rotations.
Factor rotation is motivated by the fact that factor models are not unique. To see this, let be any
orthogonal matrix. We can write the factor model as:
We can verify that the assumptions still hold under the trasform.
Varimax Rotation
Varimax rotation finds the rotation that maximizes the sum of the variances of the squared loadings, namely, find , such that
is maximized, where
and
Estimation of Factor Scores
Given the factor model:
estimate the vectors of factor scores
for each observation.
Ordinary Least Squares (OLS)
The factor scores is found by minimizing the sum of the residual squares:
Thus,
Weightead Least Squares (WLS)
The difference between WLS and OLS is that the squared residuals are divided by the specific variances . This is going to give more weight, in this estimation, to variables that have low specific variances.
The factor scores is found by minimizing the sum of the residual squares:
Thus,
Example
Data
Here is an example using package factor_analyzer. Let repeat the example using statsmodel.
The dataset contains an airline passenger satisfaction survey, which has 25976 observations and 25 columns.
Unnamed: 0 id ... Arrival Delay in Minutes satisfaction
0 0 19556 ... 44.0 satisfied
1 1 90035 ... 0.0 satisfied
2 2 12360 ... 0.0 neutral or dissatisfied
3 3 77959 ... 6.0 satisfied
4 4 36875 ... 20.0 satisfied
... ... ... ... ...
25971 25971 78463 ... 0.0 neutral or dissatisfied
25972 25972 71167 ... 0.0 satisfied
25973 25973 37675 ... 0.0 neutral or dissatisfied
25974 25974 90086 ... 0.0 satisfied
25975 25975 34799 ... 0.0 neutral or dissatisfied
[25976 rows x 25 columns]
Here is a list of the 25 columns, where the columns 8 to 21 represent customers response on a scale of 1 to 5 to a survey evaluating different aspects of the flights.
['Unnamed: 0',
'id',
'Gender',
'Customer Type',
'Age',
'Type of Travel',
'Class',
'Flight Distance',
'Inflight wifi service',
'Departure/Arrival time convenient',
'Ease of Online booking',
'Gate location',
'Food and drink',
'Online boarding',
'Seat comfort',
'Inflight entertainment',
'On-board service',
'Leg room service',
'Baggage handling',
'Checkin service',
'Inflight service',
'Cleanliness',
'Departure Delay in Minutes',
'Arrival Delay in Minutes',
'satisfaction']
The first step of any factor analysis is to look at a correlation plot of all the variables to see if any variables are useless or too correlated with others.
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
data = pd.read_csv('airline.csv',sep=',')
c = data.corr()
ax = sns.heatmap(c,square=True)
ax.figure.subplots_adjust(bottom = 0.25)
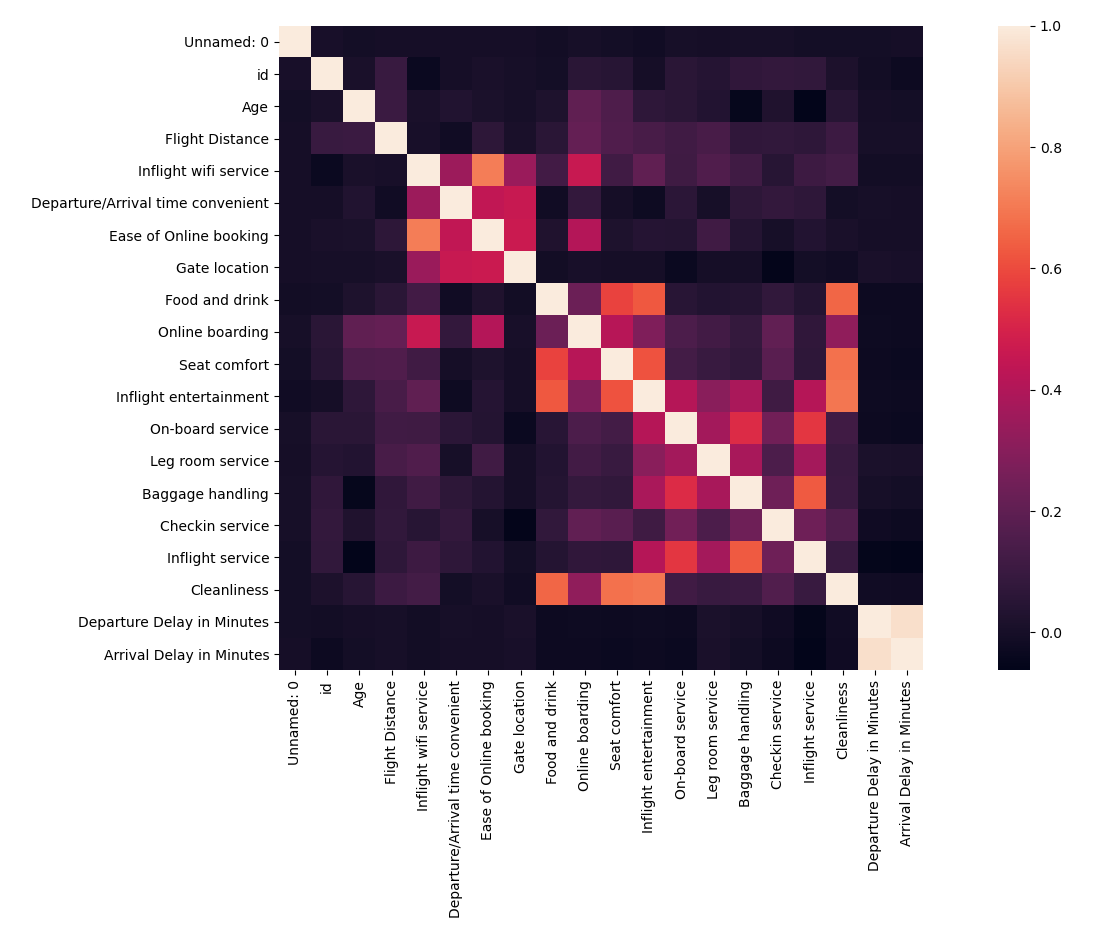
As “Departure Delay in Minutes” and the “Arrival Delay in Minutes” are highly correlated, remove one of the columns from the dataset.
data.drop(['Arrival Delay in Minutes'], axis=1, inplace=True)
Factor Analysis
class statsmodels.multivariate.factor.Factor(endog=None, n_factor=1, corr=None, method='pa', smc=True, endog_names=None, nobs=None, missing='drop')
Parameters
endog: array_like
Variables in columns, observations in rows. May be None if corr is not None.
n_factor: int
The number of factors to extract
method: str
The method to extract factors, currently must be either ‘pa’ for principal axis factor analysis or ‘ml’ for maximum likelihood estimation.
# 14 columns of the customers's responses to the survey
x =data[data.columns[8:22]]
# set the number of factor as 3
results = sm.Factor(x,3).fit()
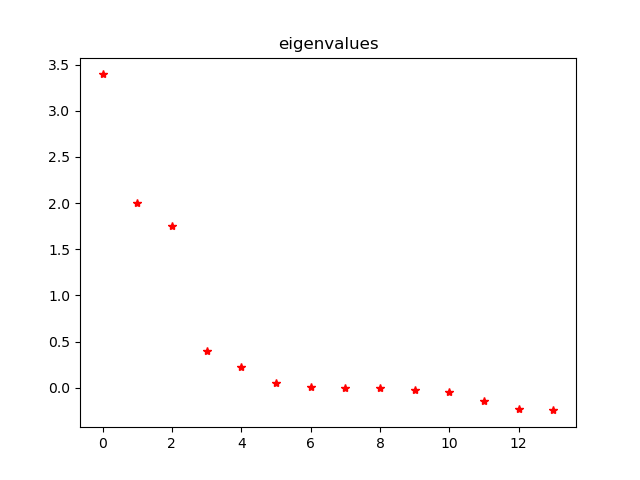
Eigenvalues:
results.eigenvals
Out[116]:
array([ 3.39287070e+00, 1.99752234e+00, 1.74684264e+00, 4.03928420e-01,
2.25980639e-01, 4.79457525e-02, 1.14410507e-02, -1.96191104e-03,
-5.44379913e-03, -2.43802755e-02, -4.67822926e-02, -1.43716548e-01,
-2.23380565e-01, -2.43630473e-01])
Communality:
results.communality
Out[124]:
array([0.62136998, 0.26925258, 0.85027123, 0.27214144, 0.57158413,
0.2980277 , 0.63998574, 0.78040315, 0.50684261, 0.25212025,
0.5932482 , 0.10115273, 0.6385212 , 0.74231474])
Unrotated loadings:
results.loadings
Out[123]:
array([[ 0.41623784, -0.65850723, 0.12035065],
[ 0.14214463, -0.4986264 , 0.0204743 ],
[ 0.30185314, -0.8578344 , 0.15256493],
[ 0.0965834 , -0.50311275, 0.09844108],
[ 0.57728164, 0.25446329, 0.41662749],
[ 0.46516202, -0.21285358, 0.19064456],
[ 0.65348712, 0.24435365, 0.3914481 ],
[ 0.84415447, 0.25585916, 0.04839901],
[ 0.48650244, 0.02597749, -0.51911767],
[ 0.36742193, -0.03771079, -0.34014596],
[ 0.48111541, 0.01022714, -0.60139137],
[ 0.27483868, 0.03324022, -0.15656155],
[ 0.49262959, 0.0194701 , -0.62885468],
[ 0.69879915, 0.29553957, 0.40822891]])
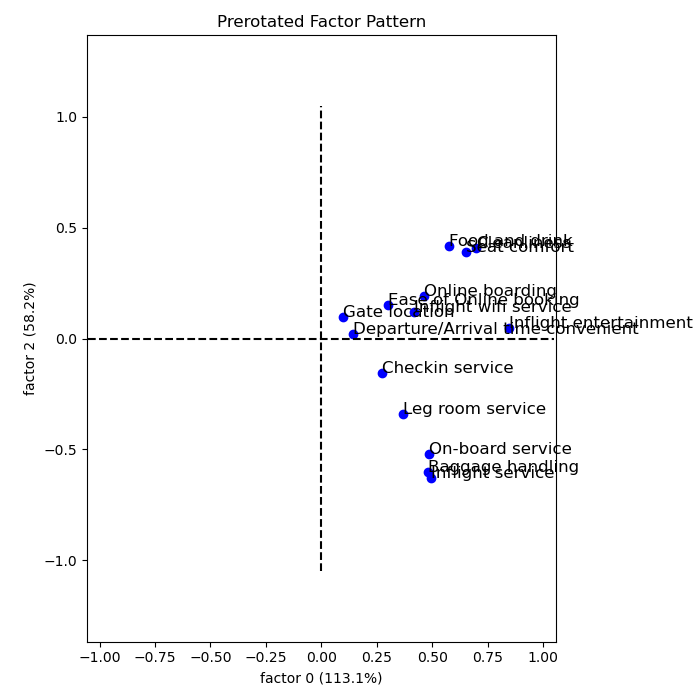
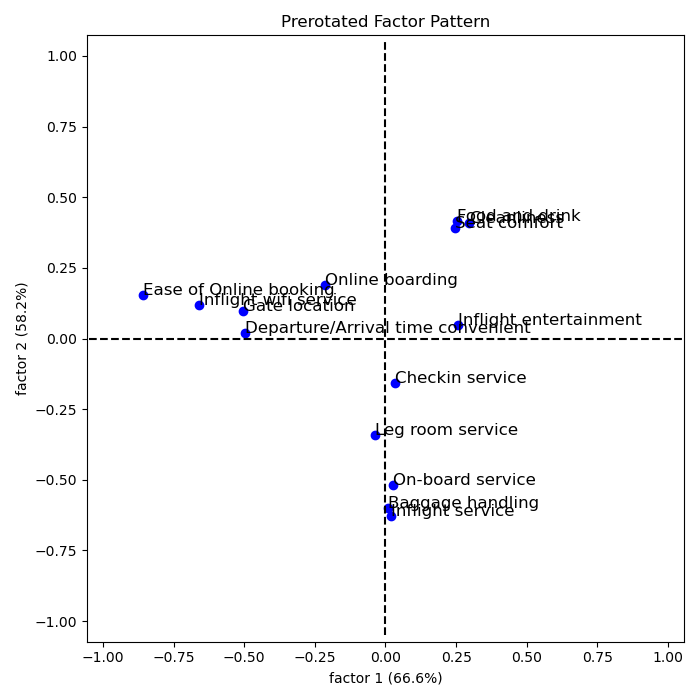
Rotated loadings:
results.rotate('varimax')
results.loadings
Out[132]:
array([[ 0.14787756, -0.76735192, -0.10331135],
[-0.05930589, -0.51341631, -0.04625023],
[ 0.001848 , -0.92203905, -0.01057408],
[-0.05756442, -0.51660793, 0.04409107],
[ 0.75467628, -0.02013316, 0.04052768],
[ 0.38346871, -0.38016993, -0.08031359],
[ 0.79818003, -0.04952743, -0.02100997],
[ 0.77983455, -0.04002052, -0.41310965],
[ 0.13074484, -0.04093086, -0.69862226],
[ 0.103968 , -0.09334843, -0.48228309],
[ 0.07949962, -0.03954792, -0.76509083],
[ 0.1492195 , -0.02964238, -0.27929841],
[ 0.07811092, -0.02983718, -0.79468839],
[ 0.86076469, -0.0192643 , -0.03205879]])
The higher a factor loading, the more important a variable is for said factor. Usually, a loading cutoff of 0.5 is used. This cutoff decides which variables belongs to which factor. Hence,
- The first factor caontains variables 5, 7, 8, 14
- The second factor contains variables 1, 2, 3, 4
- The third factor contains variables 9, 11, 13
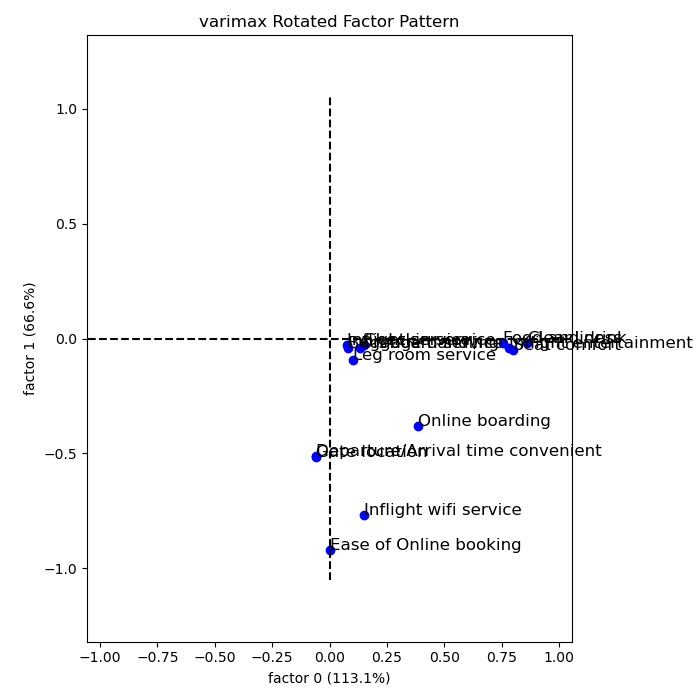
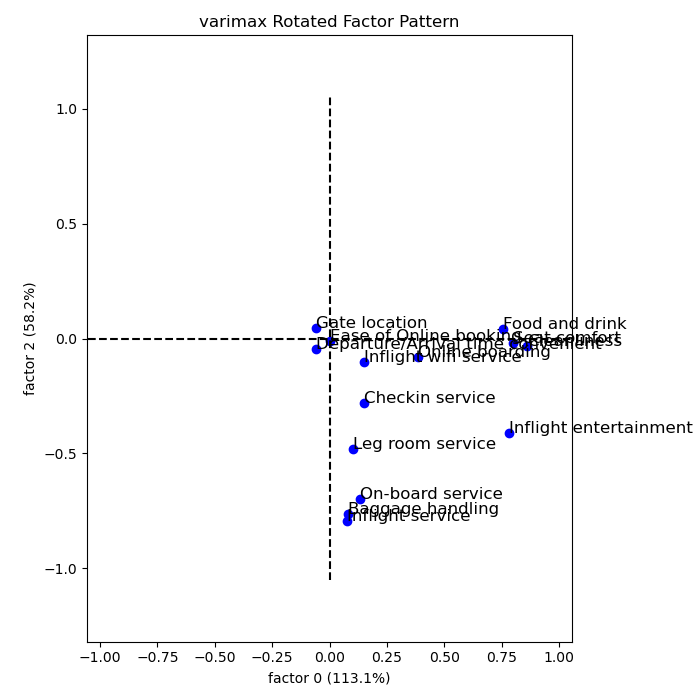
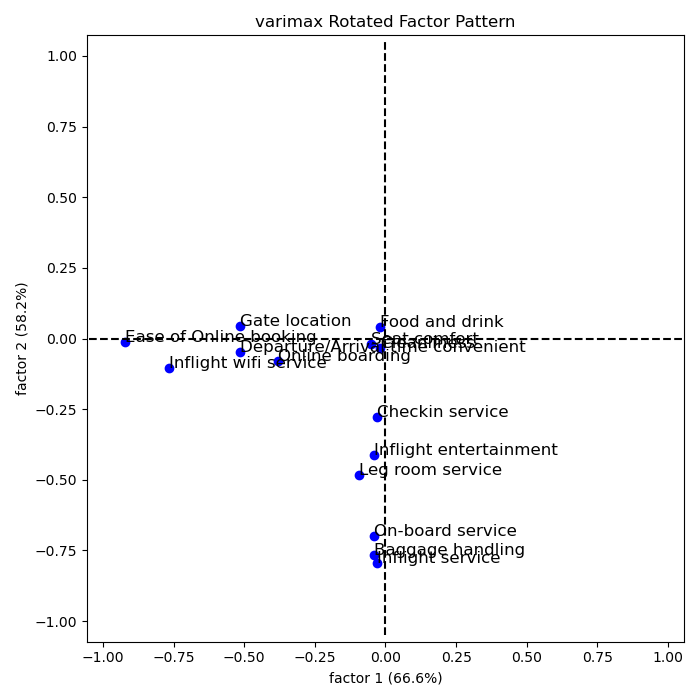
Recall that the 14 variables considered are:
['Inflight wifi service',
'Departure/Arrival time convenient',
'Ease of Online booking',
'Gate location',
'Food and drink',
'Online boarding',
'Seat comfort',
'Inflight entertainment',
'On-board service',
'Leg room service',
'Baggage handling',
'Checkin service',
'Inflight service',
'Cleanliness']
Therefore, the three factors can be explained as:
- Comfort: Food and Drink, Seat comfort, Inflight entertainment, Cleanliness
- Convenience: In flight Wifi, Departure/Arrival time convenience, Online Booking, Gate Location.
- Service: Onboard service, Baggage Handling, Inflight Service
Factor Scores
Factor scoring coefficient matrix:
results.factor_score_params()
Out[139]:
array([[ 0.01729959, -0.24417132, -0.01322147],
[-0.01900684, -0.08682621, -0.00894957],
[-0.0535145 , -0.76064598, 0.05825796],
[-0.0126019 , -0.0891501 , 0.01693065],
[ 0.22051131, 0.00780611, 0.10849854],
[ 0.05676871, -0.06106693, 0.01001721],
[ 0.26798272, 0.0022086 , 0.10067651],
[ 0.33992157, 0.02923623, -0.20999439],
[-0.03919222, 0.00858151, -0.27956411],
[-0.01631891, -0.00670421, -0.12531997],
[-0.07114288, 0.01168981, -0.37920286],
[ 0.00451548, 0.0009571 , -0.05573909],
[-0.08440538, 0.01740522, -0.44418667],
[ 0.403036 , 0.02014393, 0.14256182]])
Factor scores:
results.factor_scoring()
Out[140]:
array([[ 0.5894652 , -0.61037357, -1.52316734],
[ 1.26600869, 0.46717871, -0.06557448],
[-1.07729015, 0.62376949, 0.83407618],
...,
[-1.3706222 , 0.88601783, -0.62703381],
[ 0.50028647, -0.14472913, -0.67078162],
[-1.10915367, 0.18519492, 2.65380433]])
Principle Component Analysis
In factor analysis we model the observed variables as linear functions of the “factors.” In principal components, we create new variables that are linear combinations of the observed variables.
Notations and Model
Suppose we have a random vector with population variance matrix
Consider the linear combination
Each of these can be considered as a multiple regression without intercepts.
For
and
PCA Procedure
First Principle Component (PCA1)
The first principal component is the linear combination of x-variables that has maximum variance (among all linear combinations). It accounts for as much variation in the data as possible.
We define the coefficeients for the first component in such as way that its variance is maximized, subject to the constraint that the sum of the squared coefficients is equal to 1. Namely, select to maximize
subject to
Second Principle Component (PCA2)
The second principal component is the linear combination of x-variables that accounts for as much of the remaining variation as possible, with the constraint that the correlation between the first and second component is 0.
Namely, select to maximize
subject to
and
i-th Principle Component (PCAi)
select to maximize
subject to
and
Solution
Let denote the
eigenvalues of
with
The corresponding eigenvectors are denoted as
It turns out that the elements for these eigenvectors are the coefficients of the principal components.
The variance for the principle component is
and
By spectral decomposition theorem, we have
The idea behind PCA is to approximate the covariance matrix by tahe first components, namely
To understand this, we define the total variation of as the trace of the covariance matrix, which is the sum of the variances of the individual variables. That is
Thus, the principle component explains the following proportion of the total variation:
Therefore, the first principle component would explain the following proportion of the total variation:
Example
We use the same data as the example in factor analysis and still use the statsmodels to realize PCA algorithm.
class statsmodels.multivariate.pca.PCA(data, ncomp=None, standardize=True, demean=True, normalize=True, gls=False, weights=None, method='svd', missing=None, tol=5e-08, max_iter=1000, tol_em=5e-08, max_em_iter=100, svd_full_matrices=False)
Parameters:
data: array_like
Variables in columns, observations in rows.
ncomp: int, optional
Number of components to return. If None, returns the as many as the smaller of the number of rows or columns in data.
Data Analysis
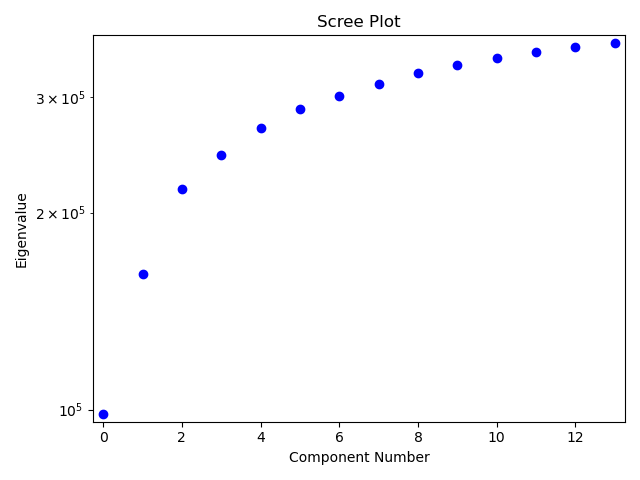
First, let examine the eigenvalues to determine how many principle components should be considered.
results = sm.PCA(x, method = 'nipals')
Eigenvalues:
results.eigenvals
Out[292]:
0 98351.323505
1 62565.929983
2 56669.672061
3 27624.149068
4 24520.057939
5 17897.417218
6 13734.114836
7 13188.024211
8 12018.743569
9 9481.617208
10 8586.135543
11 7465.813471
12 6741.359084
13 4819.642304
Name: eigenvals, dtype: float64
Below is the cumulative percentage obtained by adding the successive proportions of variation explained by each eigenvalue.
[0.2704455857730838,
0.4424888179407722,
0.5983185730459308,
0.674279209974658,
0.7417042450053434,
0.7909184020783133,
0.8286843476664585,
0.8649486581584533,
0.8979976912454709,
0.9240701570614314,
0.9476802354388428,
0.9682096622480078,
0.9867469908923509,
1.0]
And below is the scree plot of the eigenvalues, to cover 80% of the variation, we need 7 principle components.
PCA
Next, we compute the principal component scores using loadings.
results = sm.PCA(x,7,method = 'nipals')
Loadings: ncomp by nvar array of principal component loadings for constructing the factors
results.loadings
Out[316]:
comp_0 comp_1 ... comp_5 comp_6
Inflight wifi service 0.229428 0.448378 ... -0.095192 -0.222513
Departure/Arrival time convenient 0.089759 0.423246 ... -0.007206 0.600277
Ease of Online booking 0.158174 0.530357 ... -0.022008 -0.133023
Gate location 0.059700 0.430829 ... 0.175737 -0.304796
Food and drink 0.307710 -0.170796 ... 0.004112 -0.168312
Online boarding 0.277880 0.157983 ... -0.185668 0.199786
Seat comfort 0.345547 -0.159879 ... 0.066538 0.194946
Inflight entertainment 0.431140 -0.174646 ... -0.058597 -0.034484
On-board service 0.277950 -0.050805 ... -0.225194 0.414931
Leg room service 0.226631 0.001451 ... 0.840270 0.153394
Baggage handling 0.268328 -0.040025 ... -0.206501 -0.242053
Checkin service 0.178831 -0.044026 ... 0.238955 -0.294801
Inflight service 0.270867 -0.046380 ... -0.240973 -0.175362
Cleanliness 0.357168 -0.185636 ... 0.050917 -0.013267
[14 rows x 7 columns]
For PCA, loadings are the same as eigenvectors:
results.eigenvecs
Out[319]:
eigenvec_0 eigenvec_1 eigenvec_2 ... eigenvec_4 eigenvec_5 eigenvec_6
0 0.229428 0.448378 -0.069026 ... -0.226457 -0.095192 -0.222513
1 0.089759 0.423246 0.007340 ... 0.407065 -0.007206 0.600277
2 0.158174 0.530357 -0.069181 ... -0.158245 -0.022008 -0.133023
3 0.059700 0.430829 -0.060077 ... 0.152772 0.175737 -0.304796
4 0.307710 -0.170796 -0.345555 ... -0.000059 0.004112 -0.168312
5 0.277880 0.157983 -0.169104 ... -0.095087 -0.185668 0.199786
6 0.345547 -0.159879 -0.315762 ... 0.102873 0.066538 0.194946
7 0.431140 -0.174646 -0.070707 ... -0.145801 -0.058597 -0.034484
8 0.277950 -0.050805 0.386667 ... -0.003841 -0.225194 0.414931
9 0.226631 0.001451 0.299764 ... -0.322904 0.840270 0.153394
10 0.268328 -0.040025 0.429205 ... -0.005617 -0.206501 -0.242053
11 0.178831 -0.044026 0.144506 ... 0.764468 0.238955 -0.294801
12 0.270867 -0.046380 0.437012 ... -0.004266 -0.240973 -0.175362
13 0.357168 -0.185636 -0.314051 ... 0.070783 0.050917 -0.013267
[14 rows x 7 columns]
Scores (factors):
results.scores
Out[320]:
comp_0 comp_1 comp_2 comp_3 comp_4 comp_5 comp_6
0 0.008190 0.004232 0.004682 -0.005359 -0.009091 -0.001165 0.002354
1 0.004861 -0.009642 -0.002719 0.003397 -0.004955 -0.000080 0.001975
2 -0.009640 -0.001508 -0.000549 -0.001721 -0.004047 -0.008081 -0.007137
3 -0.011144 -0.010268 -0.013178 0.007958 0.003495 0.000297 -0.000113
4 -0.006631 0.001024 -0.006514 -0.002883 0.006178 0.003718 -0.007019
... ... ... ... ... ... ...
25971 0.002635 -0.003803 -0.000637 0.002205 0.002847 -0.009210 -0.003536
25972 0.008337 0.004220 0.003988 0.000708 0.003685 0.005225 -0.005247
25973 -0.004935 0.002529 0.008071 -0.008449 0.014689 0.003291 -0.001168
25974 0.004313 -0.000786 -0.000022 -0.000034 0.003890 -0.009497 -0.008091
25975 -0.013136 0.006459 -0.009547 -0.009968 0.000275 0.004297 0.005153
[25976 rows x 7 columns]
Interpretation
To interpret each component, we should compute the correlations between the original data and each principle component.
x_np = x.to_numpy()
y_np = results.factors.to_numpy()
corrM = np.corrcoef(np.transpose(x_np),np.transpose(y_np))
corrP = corrM[:14,14:]
Results:
PCA1 PCA2 PCA3 PCA4 PCA5 PCA6 PCA7
X1 array([[4.4643e-01, 6.9587e-01, -1.0195e-01, 1.9242e-01, -2.2002e-01, -7.9015e-02, -1.6180e-01],
X2 [ 1.7466e-01, 6.5686e-01, 1.0842e-02, -3.1111e-01, 3.9549e-01, -5.9812e-03, 4.3648e-01],
X3 [ 3.0778e-01, 8.2310e-01, -1.0218e-01, 1.2615e-01, -1.5375e-01, -1.8268e-02, -9.6726e-02],
X4 [ 1.1616e-01, 6.6863e-01, -8.8735e-02, -4.7870e-01, 1.4843e-01, 1.4587e-01, -2.2163e-01],
X5 [ 5.9875e-01, -2.6507e-01, -5.1040e-01, -2.1042e-01, -5.7728e-05, 3.4130e-03, -1.2239e-01],
X6 [ 5.4071e-01, 2.4519e-01, -2.4977e-01, 6.1012e-01, -9.2383e-02, -1.5412e-01, 1.4527e-01],
X7 [ 6.7237e-01, -2.4813e-01, -4.6639e-01, 1.0980e-02, 9.9949e-02, 5.5231e-02, 1.4175e-01],
X8 [ 8.3892e-01, -2.7105e-01, -1.0444e-01, -2.2680e-01, -1.4166e-01, -4.8639e-02, -2.5074e-02],
X9 [ 5.4084e-01, -7.8848e-02, 5.7112e-01, -9.8156e-03, -3.7317e-03, -1.8692e-01, 3.0171e-01],
X10 [ 4.4098e-01, 2.2526e-03, 4.4276e-01, 6.6841e-02, -3.1372e-01, 6.9747e-01, 1.1154e-01],
X11 [ 5.2212e-01, -6.2117e-02, 6.3395e-01, -1.0243e-01, -5.4570e-03, -1.7141e-01, -1.7600e-01],
X12 [ 3.4797e-01, -6.8327e-02, 2.1344e-01, 4.1332e-01, 7.4273e-01, 1.9835e-01, -2.1436e-01],
X13 [ 5.2706e-01, -7.1981e-02, 6.4548e-01, -1.2812e-01, -4.1446e-03, -2.0002e-01, -1.2751e-01],
X14 [ 6.9499e-01, -2.8810e-01, -4.6386e-01, -1.1913e-01, 6.8771e-02, 4.2264e-02, -9.6470e-03]])
Similar to factor analysis, if the correlation is greater than 0.5, we consider it as important. Thus,
- PCA1: +X5, +X6, +X7, +X8, +X9, +X11, +X13, +X14
- PCA2: +X1, +X2, +X3, +X4
- PCA3: -X5, +X9, +X11, +X13
- PCA4: +X6
- PCA5: +X12
- PCA6: +X10
- PCA7: +X2
We can also check that the covariance matrix of the principle components is an identical matrix.
corrM[14:,14:]
Out[337]:
array([[ 1.0000e+00, -4.6926e-08, -2.1001e-11, 5.2909e-17, -1.0799e-16, 3.2309e-16, -2.3419e-17],
[-4.6926e-08, 1.0000e+00, 4.7947e-08, -3.0626e-15, -3.4868e-16, -6.9389e-17, 2.2183e-16],
[-2.1001e-11, 4.7947e-08, 1.0000e+00, -3.2156e-08, -3.6600e-09, -6.1573e-13, 1.2863e-15],
[ 5.2909e-17, -3.0626e-15, -3.2156e-08, 1.0000e+00, -4.7749e-08, 6.5655e-15, -2.2985e-17],
[-1.0799e-16, -3.4868e-16, -3.6600e-09, -4.7749e-08, 1.0000e+00, -3.7023e-08, 3.4079e-13],
[ 3.2309e-16, -6.9389e-17, -6.1573e-13, 6.5655e-15, -3.7023e-08, 1.0000e+00, -3.9001e-08],
[-2.3419e-17, 2.2183e-16, 1.2863e-15, -2.2985e-17, 3.4079e-13, -3.9001e-08, 1.0000e+00]])
One can interpret these component by component. One method of deciding how many components to include is to choose only those that give unambiguous results, i.e., where no variable appears in two different columns as a significant contribution.
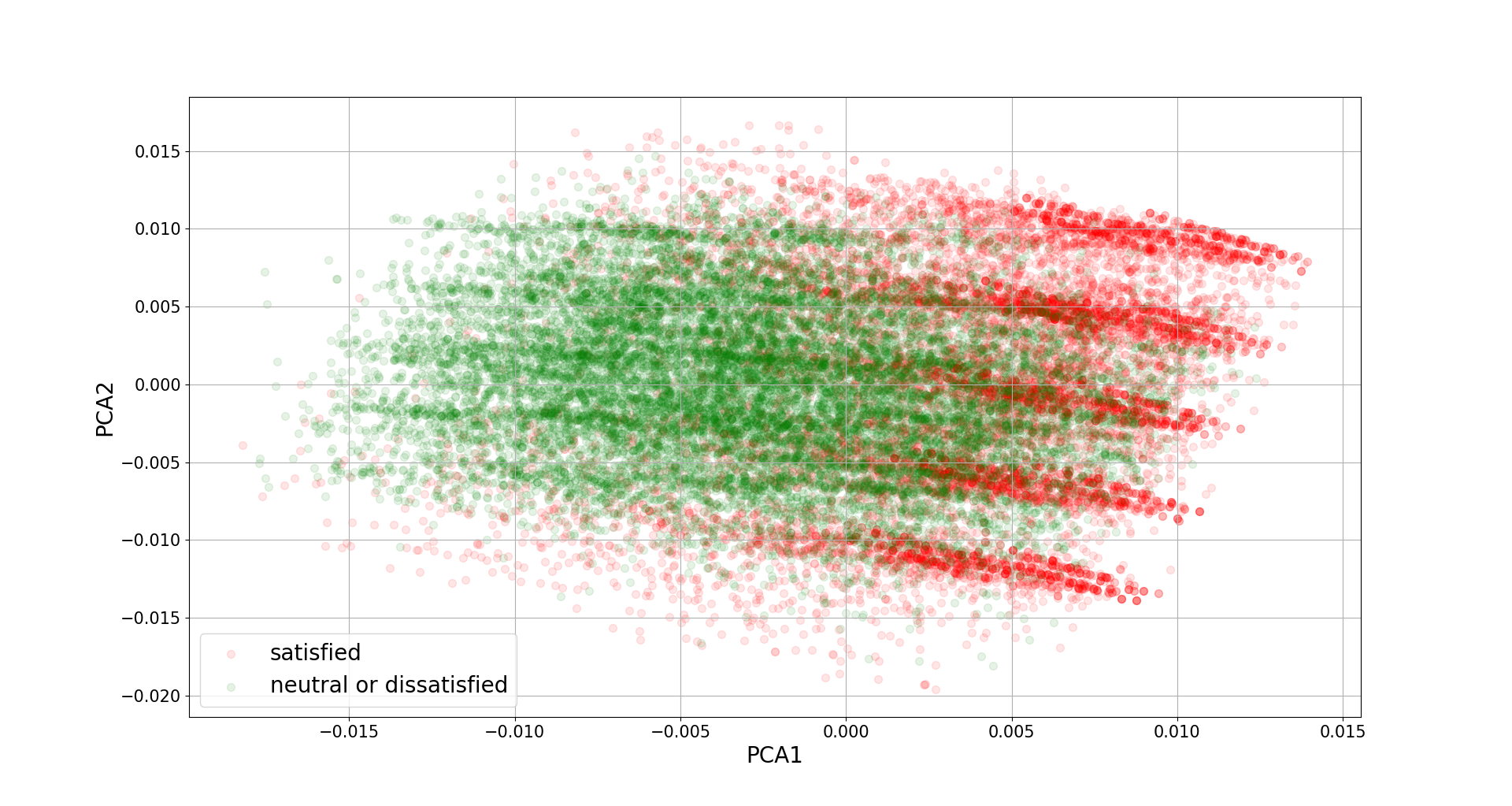
To complete the analysis we often would like to produce a scatter plot of the component scores. For example, below is a figure plotting the second component against the first one, where red dots represent satisfied and green dots represent neutral or unsatisfied.
finalDF = pd.concat([results.scores, data[['satisfaction']]], axis = 1)
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
ax.set_xlabel('PCA1', fontsize = 15)
ax.set_ylabel('PCA2', fontsize = 15)
targets = ['satisfied', 'neutral or dissatisfied']
colors = ['r', 'g']
for target, color in zip(targets,colors):
indicesToKeep = finalDF['satisfaction'] == target
ax.scatter(finalDF.loc[indicesToKeep, 'comp_0']
, finalDF.loc[indicesToKeep, 'comp_1']
, c = color
, s = 50, alpha = 0.1)
ax.legend(targets)
ax.grid()
For more examples, here is an example using sklearn and the Iris dataset.
Table of Contents
- Probability vs Statistics
- Shakespear’s New Poem
- Some Common Discrete Distributions
- Some Common Continuous Distributions
- Statistical Quantities
- Order Statistics
- Multivariate Normal Distributions
- Conditional Distributions and Expectation
- Problem Set [01] - Probabilities
- Parameter Point Estimation
- Evaluation of Point Estimation
- Parameter Interval Estimation
- Problem Set [02] - Parameter Estimation
- Parameter Hypothesis Test
- t Test
- Chi-Squared Test
- Analysis of Variance
- Summary of Statistical Tests
- Python [01] - Data Representation
- Python [02] - t Test & F Test
- Python [03] - Chi-Squared Test
- Experimental Design
- Monte Carlo
- Variance Reducing Techniques
- From Uniform to General Distributions
- Problem Set [03] - Monte Carlo
- Unitary Regression Model
- Multiple Regression Model
- Factor and Principle Component Analysis
- Clustering Analysis
- Summary

Comments