Statistics [30]: Clustering Analysis
Published:
Clustering analysis is the task of grouping a set of objects in such a way that objects in the same group are more similar to each other than to those in other groups.
Measurement of Association
Metrics
Minkowski Distance
When , it is Absolute Distance
When , it is Euclidean Distance
When , it is Chebysheve Distance
Standardized Euclidean Distance
Let the covariance matrix of be
, which is
Standardized Euclidean distance:
Mahalanobis Distance
If are independent, namely, the covariance matrix
is a diagonal matrix, then Mahalanobis distance becomes standardized Euclidean distance.
Canberra Metric
Czekanowski Coefficient
Properties of Metrics
- Symmetry:
- Positivity:
- Identity:
- Triangle Inequality:
Agglomerative Clustering
Basic Ideas
In agglomerative hierarchical algorithms, we start by defining each data point as a cluster. Then, the two closest clusters are combined into a new cluster. In each subsequent step, two existing clusters are merged into a single cluster.
There are several methods for measuering association between the clusters:
- Single Linkage:
- Complete Linkage:
- Average Linkage:
- Centroid Method:
- Ward’s Method: ANOVA based approach
Example
Below is an example using package sklearn and the Iris dataset.
class sklearn.cluster.AgglomerativeClustering(n_clusters=2, *, affinity='euclidean', memory=None, connectivity=None, compute_full_tree='auto', linkage='ward', distance_threshold=None, compute_distances=False)
Data
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import normalize
from sklearn.cluster import AgglomerativeClustering
from sklearn.datasets import load_iris
# load data
iris = load_iris()
data = iris.data
data[:5]
# normalize data
data_scaled = normalize(data)
data_scaled = pd.DataFrame(data_scaled, columns=['sepalL','sepalW','petalL','petalW'])
data_scaled.head()
# Before normalization
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2]])
# After nromalization
sepalL sepalW petalL petalW
0 0.803773 0.551609 0.220644 0.031521
1 0.828133 0.507020 0.236609 0.033801
2 0.805333 0.548312 0.222752 0.034269
3 0.800030 0.539151 0.260879 0.034784
4 0.790965 0.569495 0.221470 0.031639
Draw the dendrogram to help us decide the number of clusters:
from scipy.cluster.hierarchy import dendrogram,linkage
plt.figure()
plt.title("Dendrograms")
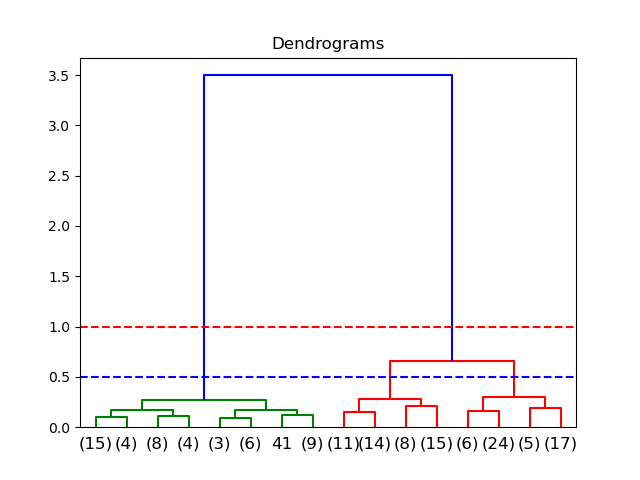
dend = dendrogram(linkage(data_scaled, method='ward'))
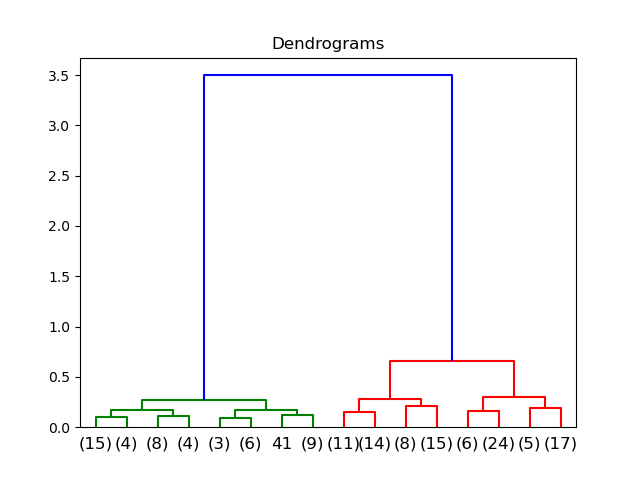
The x-axis contains the samples and y-axis represents the distance between these samples. If the threshold is 1.0, there will be two clusters; if the threshold is 0.5, there will be 3 clusters.
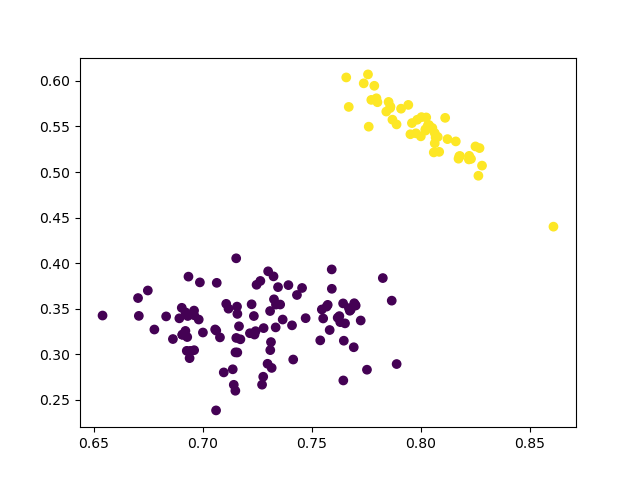
2 Clusters
First, let’s see the results of two clusters.
cluster = AgglomerativeClustering(n_clusters=2, affinity='euclidean', linkage='ward')
cluster.fit_predict(data_scaled)
array([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int64)
Visualization:
plt.figure()
plt.scatter(data_scaled['sepalL'], data_scaled['sepalW'], c=cluster.labels_)
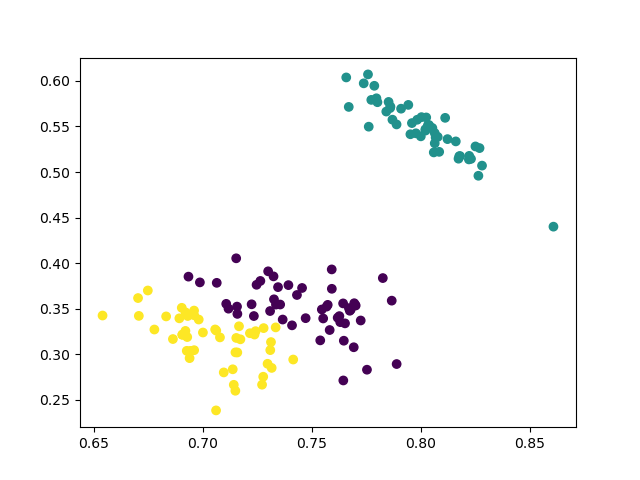
3 Clusters
cluster = AgglomerativeClustering(n_clusters=3, affinity='euclidean', linkage='ward')
cluster.labels_
array([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
0, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 0, 2, 2, 2, 0,
2, 2, 2, 2, 2, 2, 0, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2], dtype=int64)
Visualization:
plt.figure()
plt.scatter(data_scaled['sepalL'], data_scaled['sepalW'], c=cluster.labels_)
Comparison of Different Linkage Methods
Single
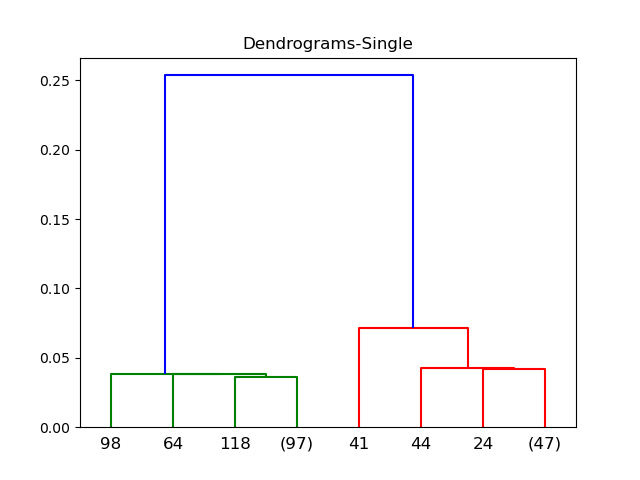
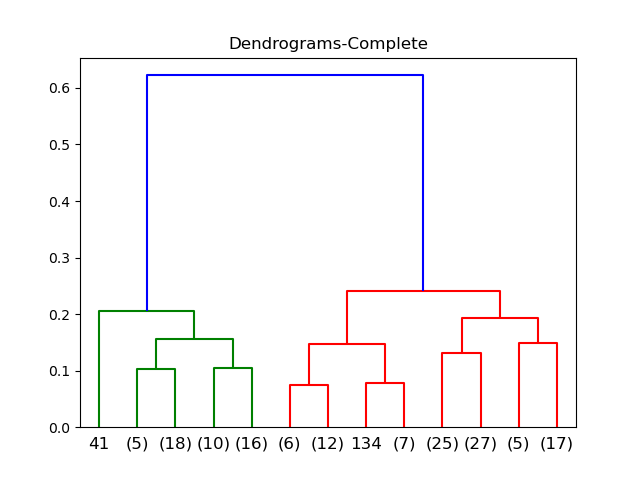
Complete
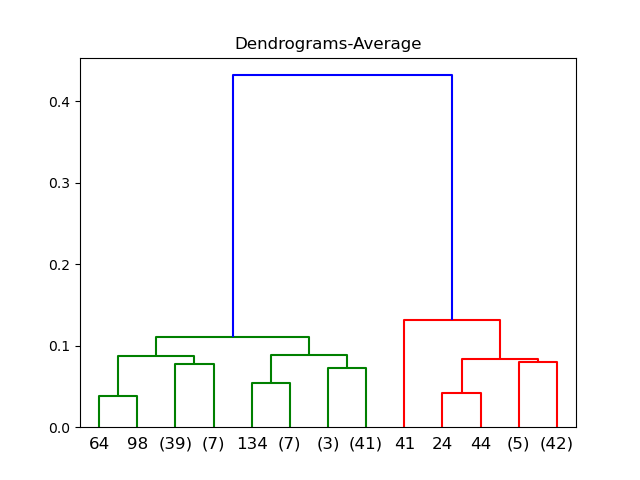
Average
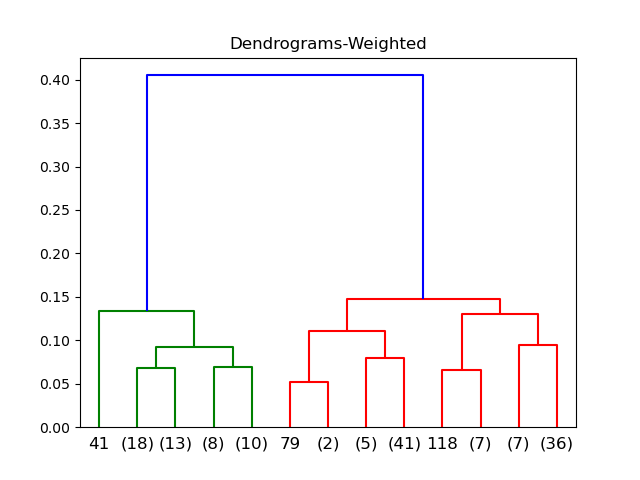
Weighted
Centroid
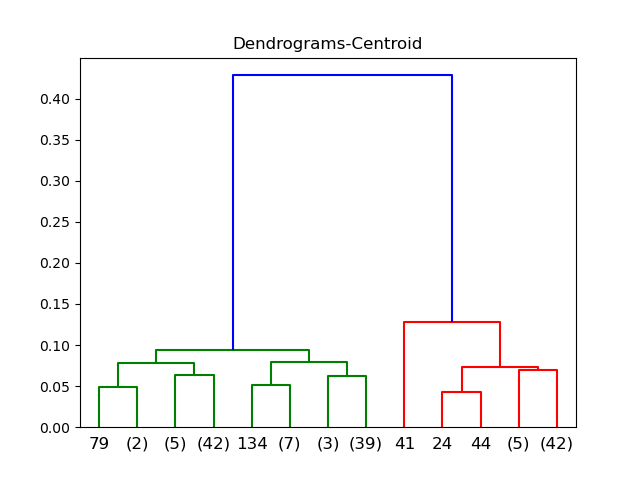
Median
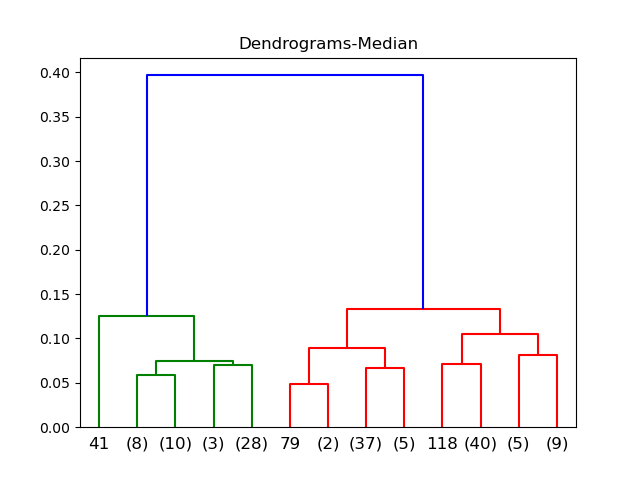
Ward
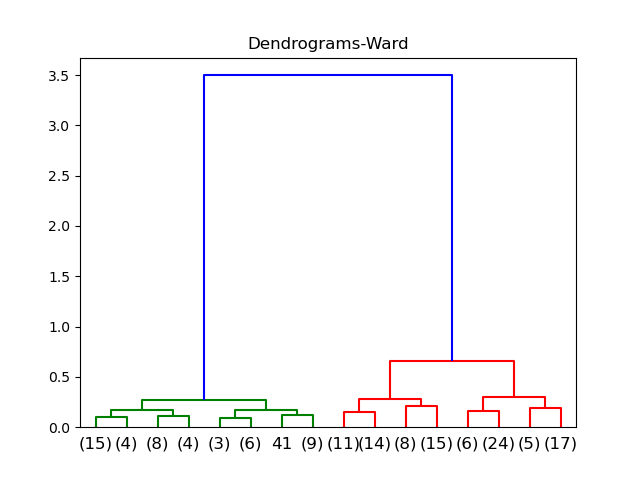
ANOVA
After obtaining the clusters, ANOVA test can be done for each of the plants, comparing the means across clusters. If the value is small, we may conclude that there are significant differences across clusters.
K-Means CLustering
Procedures
- Partition the items into
initial clusters.
- Loop thorugh the items and assign each item to the cluster whose center is closest. (Accordingly, the centroids of the clusters receiving and losing the item should be recalculated.)
- Repeat 2 until no more assignments.
Example
We repeat the above example using the Iris dataset.
from sklearn.cluster import KMeans
kmeans2 = KMeans(n_clusters=2, random_state=0).fit(data_scaled)
kmeans3 = KMeans(n_clusters=3, random_state=0).fit(data_scaled)
kmeans4 = KMeans(n_clusters=4, random_state=0).fit(data_scaled)
plt.figure()
plt.scatter(data_scaled['sepalL'], data_scaled['sepalW'], c=kmeans2.labels_)
plt.figure()
plt.scatter(data_scaled['sepalL'], data_scaled['sepalW'], c=kmeans3.labels_)
plt.figure()
plt.scatter(data_scaled['sepalL'], data_scaled['sepalW'], c=kmeans4.labels_)
2 clusters
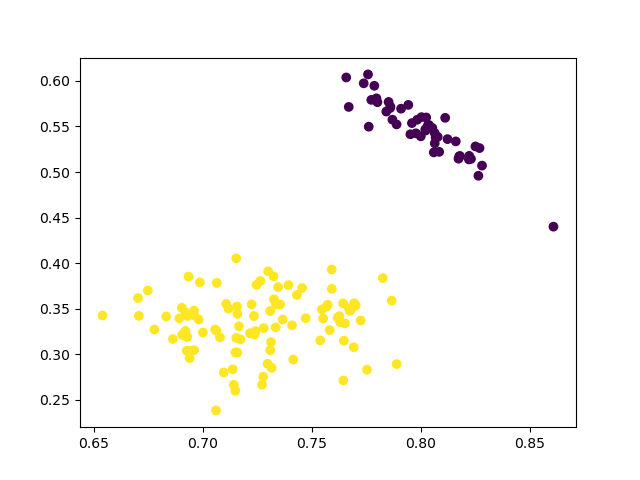
3 clusters
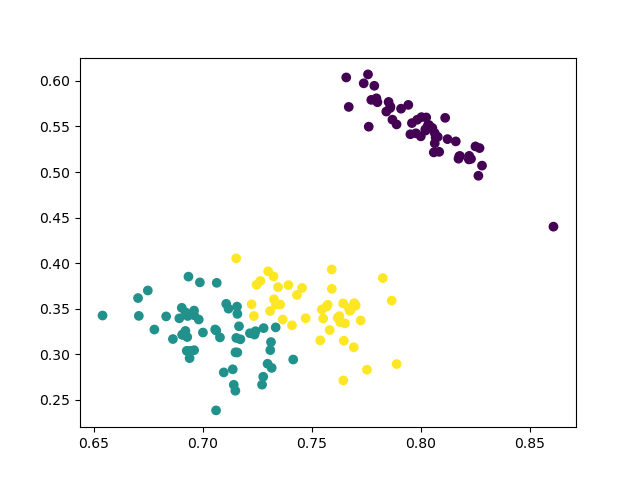
4 clusters
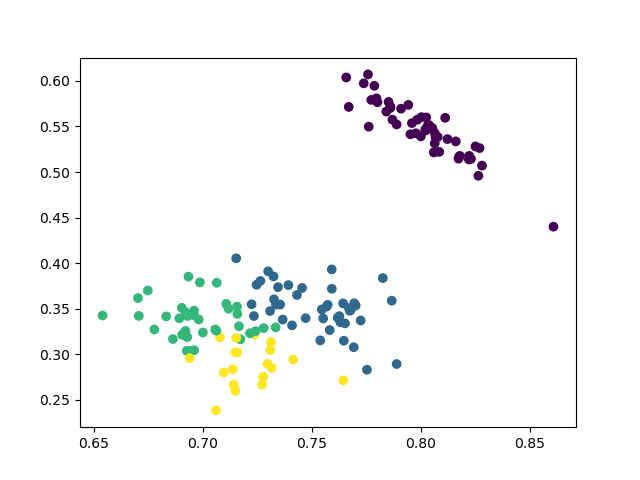
The results are similiar to the agglomerative clustering and we can also run ANOVA test to compare the means of diffearent clusters.
Table of Contents
- Probability vs Statistics
- Shakespear’s New Poem
- Some Common Discrete Distributions
- Some Common Continuous Distributions
- Statistical Quantities
- Order Statistics
- Multivariate Normal Distributions
- Conditional Distributions and Expectation
- Problem Set [01] - Probabilities
- Parameter Point Estimation
- Evaluation of Point Estimation
- Parameter Interval Estimation
- Problem Set [02] - Parameter Estimation
- Parameter Hypothesis Test
- t Test
- Chi-Squared Test
- Analysis of Variance
- Summary of Statistical Tests
- Python [01] - Data Representation
- Python [02] - t Test & F Test
- Python [03] - Chi-Squared Test
- Experimental Design
- Monte Carlo
- Variance Reducing Techniques
- From Uniform to General Distributions
- Problem Set [03] - Monte Carlo
- Unitary Regression Model
- Multiple Regression Model
- Factor and Principle Component Analysis
- Clustering Analysis
- Summary

Comments